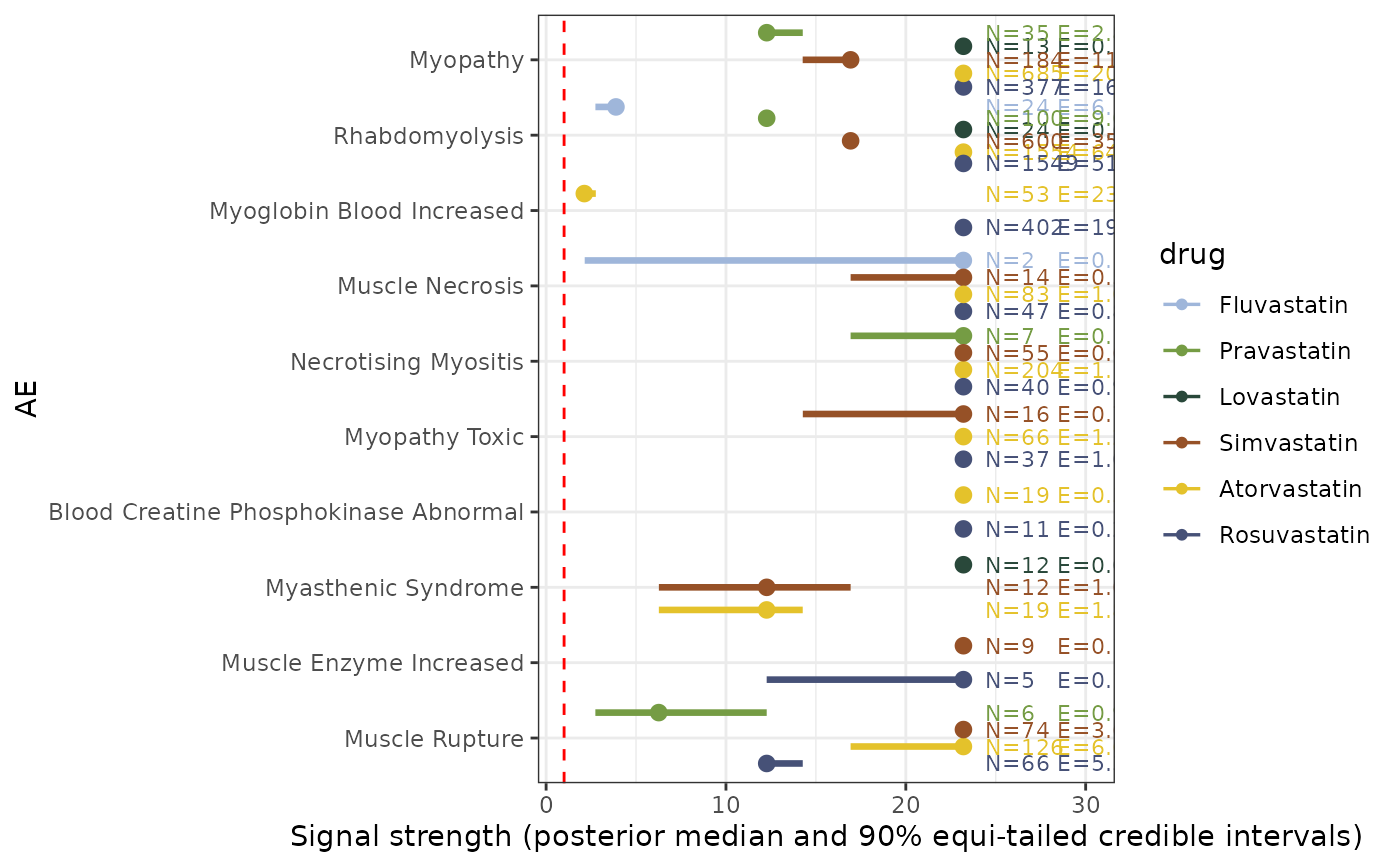
Generate an eyeplot showing the distribution of posterior draws for selected drugs and adverse events
Source:R/pvEBayes_object_S3_methods.R
eyeplot_pvEBayes.RdThis function creates an eyeplot to visualize the posterior distributions of \(\lambda_{ij}\) for selected AEs and drugs. The plot displays posterior median, 90 percent credible interval for each selected AE-drug combination.
Usage
eyeplot_pvEBayes(
x,
num_top_AEs = 10,
num_top_drugs = 8,
specified_AEs = NULL,
specified_drugs = NULL,
N_threshold = 1,
text_shift = 4,
x_lim_scalar = 1.3,
text_size = 3,
log_scale = FALSE
)Arguments
- x
a
pvEBayesobject, which is the output of the function pvEBayes or pvEBayes_tune.- num_top_AEs
a number of most significant AEs appearing in the plot. Default to 10.
- num_top_drugs
a number of most significant drugs appearing in the plot. Default to 7.
- specified_AEs
a vector of AE names that are specified to appear in the plot. If a vector of AEs is given, argument num_top_AEs will be ignored.
- specified_drugs
a vector of drug names that are specified to appear in the plot. If a vector of drugs is given, argument num_top_drugs will be ignored.
- N_threshold
a integer greater than 0. Any AE-drug combination with observation smaller than N_threshold will be filtered out.
- text_shift
numeric. Controls the relative position of text labels, (e.g., "N = 1", "E = 2"). A larger value shifts the "E = 2" further away from its original position.
- x_lim_scalar
numeric. An x-axis range scalar that ensures text labels are appropriately included in the plot.
- text_size
numeric. Controls the size of text labels, (e.g., "N = 1", "E = 2").
- log_scale
logical. If TRUE, the eye plot displays the posterior distribution of \(\log(\lambda_{ij})\) for the selected AEs and drugs.